Data conversion¶
Each MS instrument vendor has one or more formats for storing the acquired data.
Converting these data into an open format (preferably mzML) is the very first step
when you want to work with open-source mass spectrometry software. A freely available conversion tool is MSConvert, which is part of a ProteoWizard installation. All files
used in this tutorial have already been converted to mzML by us, so you do not need
to perform the data conversion yourself. However, we provide a small raw file so you
can try the important step of raw data conversion for yourself.
Note
The OpenMS installation package for Windows automatically installs ProteoWizard, so you do not need to download and install it separately. Due to restrictions from the instrument vendors, file format conversion for most formats is only possible on Windows systems. In practice, performing the conversion to mzML on the acquisition PC connected to the instrument is usually the most convenient option.
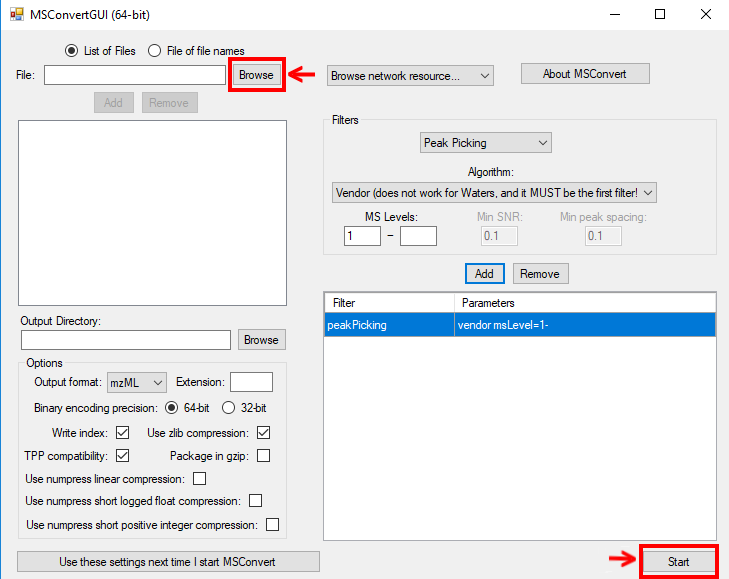
To convert raw data to mzML using ProteoWizard you can either use MSConvertGUI (a
graphical user interface) or msconvert (a simple command line tool).
|
|---|
Figure 1: |
Both tools are available in:
You can find a small RAW file on the USB stick
MSConvertGUI¶
MSConvertGUI (see Fig. 1) exposes the main parameters for data conversion in a convenient graphical user interface.
msconvert¶
The msconvert command line tool has no graphical user interface but offers more options than the application MSConvertGUI. Additionally, since it can be used within a batch script, it allows converting large numbers of files and can be much more easily automatized.
To convert and pick the file raw_data_file.RAW you may write:
msconvert raw_data_file.RAW --filter "peakPicking true 1-"
in your command line.
|
|---|
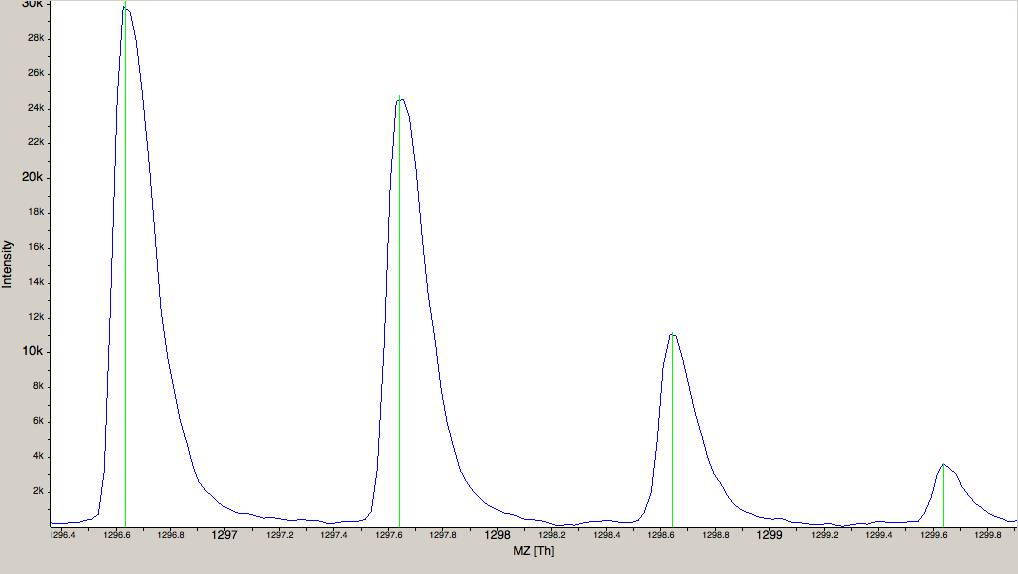
Figure 2: The amount of data in a spectra is reduced by peak picking. Here a profile spectrum (blue) is converted to centroided data (green). Most algorithms from this point on will work with centroided data. |
To convert all RAW files in a folder may write:
msconvert *.RAW -o my_output_dir
Note
To display all options you may type msconvert --help . Additional information is available on the ProteoWizard web page.
ThermoRawFileParser¶
Recently the open-source platform independent ThermoRawFileParser tool has been developed. While Proteowizard and MSConvert are only available for Windows systems this new tool allows to also convert raw data on Mac or Linux.
Note
To learn more about the ThermoRawFileParser and how to use it in
KNIME see Minimal Workflow.