TOPP Tools¶
TOPP - The OpenMS Pipeline is a set of tools for the analysis of HPLC-MS data. These tools can be either:
Applied individually using OpenMS graphical applications.
Applied in sequence as a workflow using a workflow editor such as KNIME, Nextflow or Galaxy.
Before you choose one of the above options, there are few concepts that need to be understood.
File formats¶
OpenMS only accepts files in certain formats, including but not limited to:
mzML: The HUPO-PSI standard format for mass spectrometry data.
featureXML: The OpenMS format for quantitation results.
consensusXML: The OpenMS format for grouping features in one map or across several maps.
idXML: The OpenMS format for protein and peptide identification.
Documented schemas of the OpenMS formats can be found here.
If your data is not in the above formats, you may need to use a file conversion TOPP tool.
Command Line Interface¶
Command line calls will depend on the TOPP tools used, as each TOPP tool has its own set of parameters. However, the following arguments are typically used:
-inSpecify an input file in the command line using the
-inargument. The input file should be in a supported format. If not, use the file converter to convert the file to one of the supported formats. For more information, view the file handling documentation.-outSpecify an output file in the command line using the
-outargument.-iniSpecify an INI file in the command line using the
-iniargument. TOPP uses INI files to set parameters specific to the command line tool being called.-write_iniCreate an INI file using the
-write_inifile argument. Create an INI file with this call:<insert TOPP tool> -write_ini <insert output INI File>If you want a visual tool to assist setting parameters, use the INIFileEditor, an application provided when you download OpenMS. Otherwise, you can set the parameters from the command line.-helpGet information about basic options related to the tool using the -help parameter. For more advanced options (algorithmic parameters), use
--help.--helpGet detailed information about algorithmic parameters using the
--helpparameter.
Many (but not all) command line calls will have the following structure:
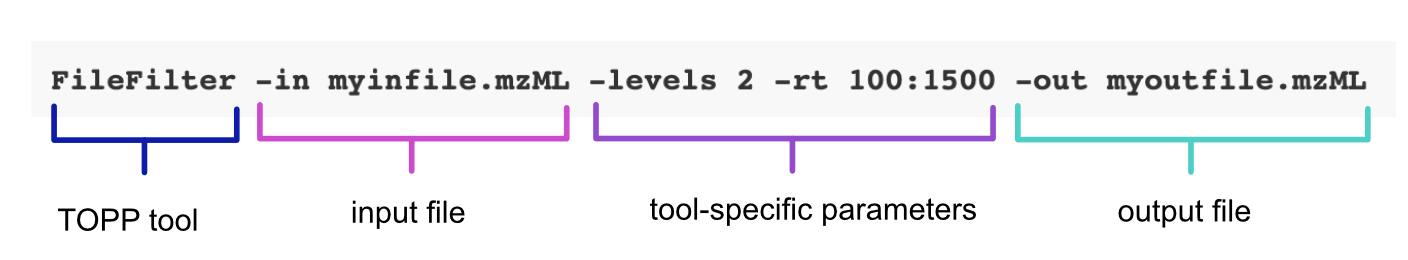
<insert TOPP tool> -in <insert input mzML file> -out <insert output mzML file> -ini <insert INI file>
The following command line call uses the FileFilter tool to extract data from an mzML file. Note, that this call directly specifies the tool-specific parameters and doesn’t rely on an INI file:
TOPP INI files¶
TOPP INI files are XML-based files with an .ini extension. OpenMS uses TOPP INI files to set parameters for one or more TOPP tools. Here is an example of a TOPP INI file:
<PARAMETERS>
<NODE name="FileFilter">
<NODE name="1">
<ITEM name="rt" value="0:1200" type="string"/>
</NODE>
<NODE name="2">
<ITEM name="mz" value="700:1000" type="string"/>
</NODE>
</NODE>
<NODE name="common">
<NODE name="FileFilter">
<ITEM name="rt" value=":" type="string"/>
<ITEM name="mz" value=":" type="string"/>
</NODE>
<ITEM name="debug" value="2" type="int"/>
</NODE>
</PARAMETERS>
Features, feature maps and featureXML files¶
An LC-MS feature is a construct in OpenMS that is used to describe a 2D peak caused by an analyte interacting with the stationary phase. Each feature contains the following metadata: an id, retention time, mass-to-charge ratio, intensity, overall quality and one or more convex hulls.
A feature map is a container for features. One feature map can contain many features.
A featureXML file is an XML based file which contains one feature map.
FeatureXML files can be created from mzML files using OpenMS’s feature detection algorithms.
Consensus feature, consensus maps, consensusXML files¶
A consensus feature is a special type of LC-MS feature that is quantified across multiple experiments. A consensus feature is formed by linking or grouping features with similar mass-to-charge ratios and intensities from various experiment runs. Each consensus feature references the features used to form the consensus feature.
Similar to a feature map, a consensus map is a container for consensus features. One consensus map can contain many consensus features.
ConsensusXML files can be created from featureXML files using OpenMS’s feature grouping algorithms.
Types of TOPP Tools¶
The following tools are offered:
File conversion
TOPP file conversion tools can be used to convert files into a supported format.
File handling
TOPP file handling tools are largely used to extract or merge data. For more information, view the File handling.
Centroiding
The conversion of the “raw” ion count data acquired by the machine into peak lists for further processing is usually called peak picking or centroiding. The choice of the algorithm should mainly depend on the resolution of the data. OpenMS provides different algorithms for centroiding depending on the resolution of the data. For more information, view the Picking peaks section.
Spectrum processing
A number of spectrum processing tools are available. These include peak filtering and peak normalization tools, as well as other miscellaneous tools.
Mass correction and calibration
To ensure that your data is sound, OpenMS have provided a number of mass correction and calibration tools. The types of tools used will depend on the type of equipment you have employed. For more information, view the Calibration section.
Spectrum clustering
Spectrum clustering is the grouping of spectra that have many peaks in common. OpenMS provides tools for spectrum clustering to identify molecules in large datasets more efficiently.
Map alignment
When looking to identify molecules, it is common to run multiple experiments, where each experiment produces a set of data. In OpenMS, every set of data is represented by a feature map. Before combining feature maps to create a consensus map, it is advised to use OpenMS’s map alignment tools so that all your datasets are comparable and based on a common retention time axis. For more information, view the Map alignment section.
Feature linking
OpenMS provides a number of algorithms for feature grouping or linking. For more information, view the Feature grouping section.
Quantitation
A number of tools are available that allow for the identification and quantification of features. The tools you use will depend on the type of mass spectrometry experiment you have set up, and the type of molecules you wish to identify. For more information, view the Feature detection section.
Protein/Peptide identification
Protein/Peptide processing
Targeted experiments and OpenSWATH
Cross-linking
Cross-linking is a technique where substances are chemically treated to create covalent bonds between different molecules. The strength of the covalent bonds can be quantified to indicate the proximity of certain molecules within a 3D structure.
Quality control
OpenMS provides tools to measure the quality of LC-MS data. For more information, view the Quality control section.
For the full list of TOPP tools, visit the API reference website.