Feature Grouping¶
In order to quantify differences across maps (label-free) or within a map (isotope-labeled), groups of corresponding features have to be found. The FeatureLinker TOPP tools support both approaches. These groups are represented by consensus features, which contain information about the constituting features in the maps as well as average position, intensity, and charge.
Isotope-labeled quantitation¶
To differentially quantify the features of an isotope-labeled HPLC-MS map, follow the listed steps:
The first step in this pipeline is to find the features of the HPLC-MS map. The FeatureFinder applications calculate the features from profile data or centroided data.
In the second step, the labeled pairs (e.g. light/heavy labels of ICAT) are determined by the FeatureLinkerLabeled application. FeatureLinkerLabeled first determines all possible pairs according to a given optimal shift and deviations in RT and m/z. Then it resolves ambiguous pairs using a greedy-algorithm that prefers pairs with a higher score. The score of a pair is the product of:
feature quality of feature 1
feature quality of feature 2
quality measure for the shift (how near is it to the optimal shift)

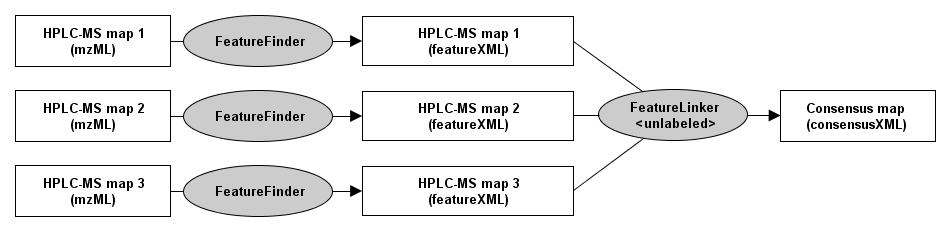
Label-free quantitation¶
To differentially quantify the features of two or more label-free HPLC-MS map.
Tip
This algorithm assumes that the retention time axes of all input maps are very similar. To correct for retention time distortions, please have a look at Map alignment.